Phylogenetic Analysis of Reticulate Software Evolution (MSR2023) Phylogenetic Analysis of Reticulate Software Evolution (MSR2023)
2023.07.13
AST differencingphylogenetic networksoftware product linesoftware evolution
2023.07.13
AST differencingphylogenetic networksoftware product linesoftware evolution

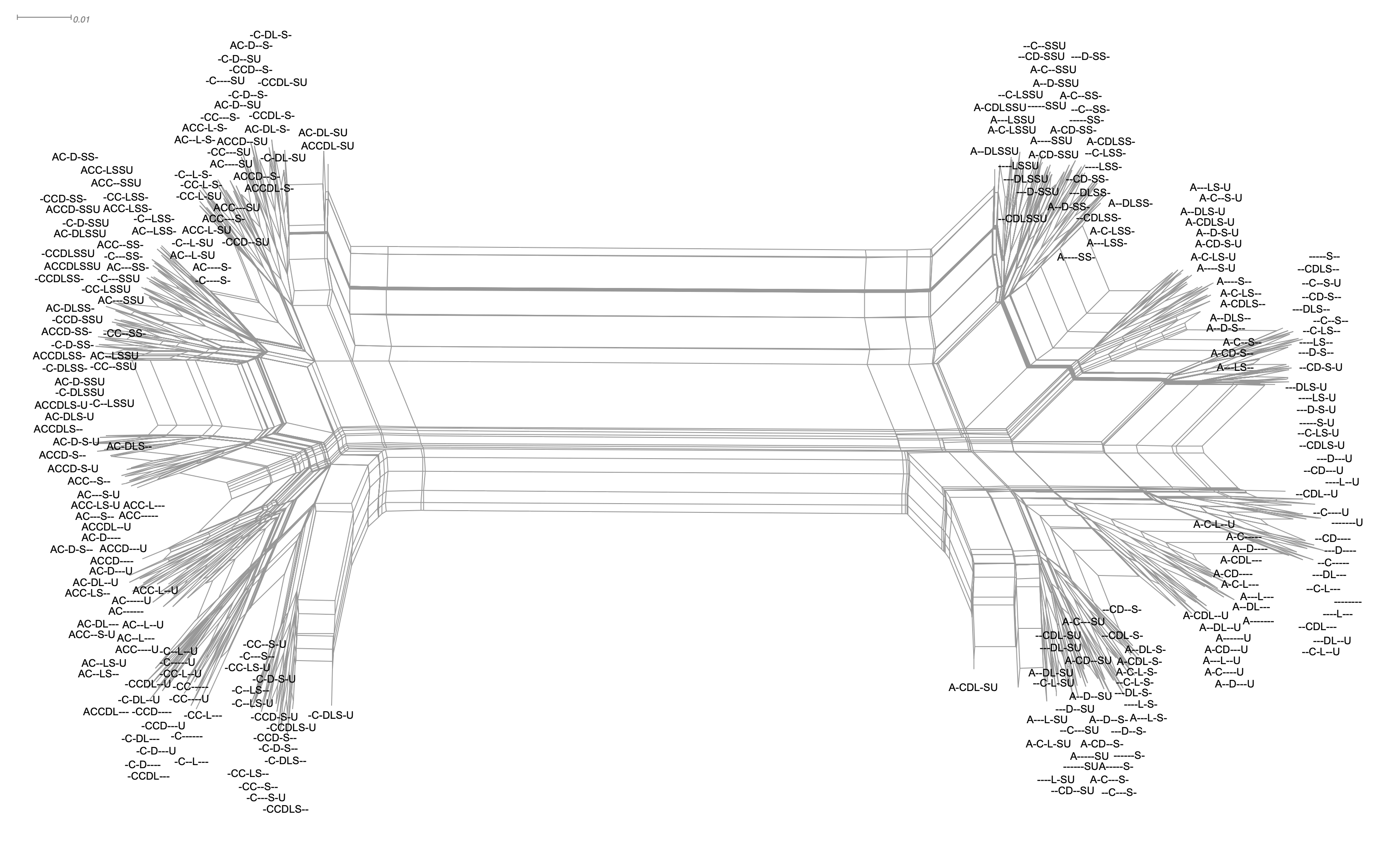
In this paper, we apply techniques from phylogenetics for uncovering evolutionary dependencies among software versions. Phylogenetics is a part of computational molecular biology that addresses the inference of evolution among organisms based on differences/similarities in DNA sequences and morphology. We apply a tree differencing technique to abstract syntax trees to calculate a distance matrix, which is then used by a distance-based phylogenetic algorithm to infer an evolution network. This allows us to identify merging and branching among versions without manually looking into the details of the source code. Experiments on ancient/old versions of the Emacs editor and the open source 3D printer firmware show that we not only can reproduce the evolution of the software but also can identify code import/merging across different lineages. We also discuss how the techniques are used to identify the feature models among software variations. To the best of our knowledge, this paper is the first to report on a reticulate phylogenetic analysis of the software and may offer a useful method for gaining information on the evolution of the software.
Akira Mori and Masatomo Hashimoto. Phylogenetic Analysis of Reticulate Software Evolution. In Proceedings of the 20th International Conference on Mining Software Repositories (MSR 2023), pp. 498-510, 2023.
A replication package of the experiments will be found here.